A longer edition of Polars in Aggregate. We have been implementing a wide range of improvements in the past weeks. This edition covers support for streaming outer joins, enhanced zero-copy NumPy interop, COUNT(*) fast path, very fast boolean filters and ternary operations, and more. Also don’t forget to have a look at the new integrations and give them a try.
Time to dive in.
Note: All benchmarks in this edition were executed on an m6i.4xlarge (vCPU 16, RAM 64GB) instance type on AWS. The scripts are available as Github Gists. You can find the links to the gists at the end of this blog post, so that you can run the benchmarks yourself as well.
Streaming outer join (#14828)
Our streaming engine now supports full outer joins. This enables more users to get their streaming queries finished. Coming time we are working on preliminary work in our optimizer and after that many more streaming operations may be expected.
Boolean filter speed up (#14746)
We have improved the performance of our filters, especially for boolean values. The new implementations are branchless and use SIMD where appropriate.
For example, suppose you have a dataframe of visitors in your amusement park with schema {"age": pl.UInt8, "height": pl Float32, "rode_rollercoaster": pl.Boolean}, where the ages just happen to be uniformly distributed integers 0-99.
We can test the performance of our filter by an example query of getting the mean height of people younger than a given age. In NumPy you might write this as np.mean(heights[ages < cutoff]), in Polars as df.select(pl.col.height.filter(pl.col.age < cutoff).mean()).
This gives the following plot, again showing the times over different selectivity percentages:
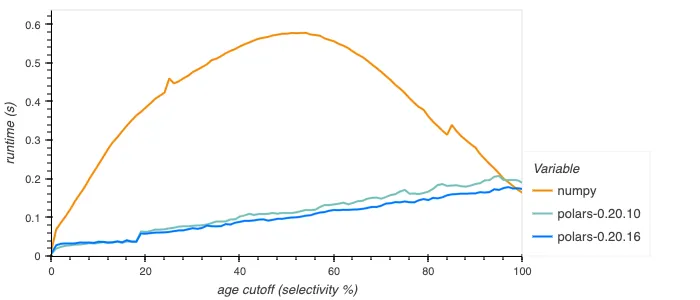
Note that NumPy’s implementation isn’t branchless: the CPU executes different instructions for the true and false cases, costing a lot of performance if the predicate is unpredictable, leading to a lot of branch mispredicts.
Especially the boolean filter performance has improved, for example repeating the above query but getting the percentage of people under a certain age that rode rollercoasters:
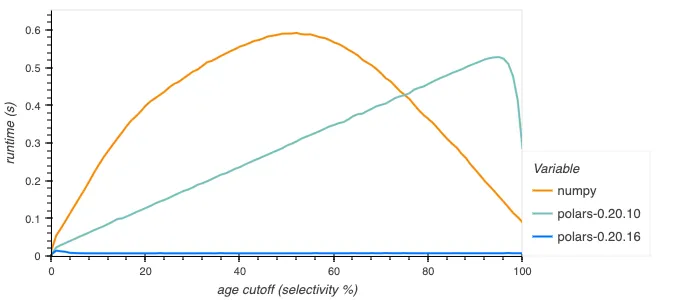
This is due to the new implementation being able to use special instructions like Intel BMI2’s PEXT or carryless multiplication on Arm to handle 64 bits at once, whereas the old implementation looped over each set bit separately.
When-then-otherwise performance (#15089)
The performance of when/then/otherwise has been improved, for example when computing the mean height of people, assuming people below a certain age are all 1 meter tall. This is df.select(pl.when(pl.col.age < cutoff).then(1).otherwise(pl.col.height).mean()) in Polars or np.mean(np.where(ages < cutoff, 1.0, heights)) in NumPy:
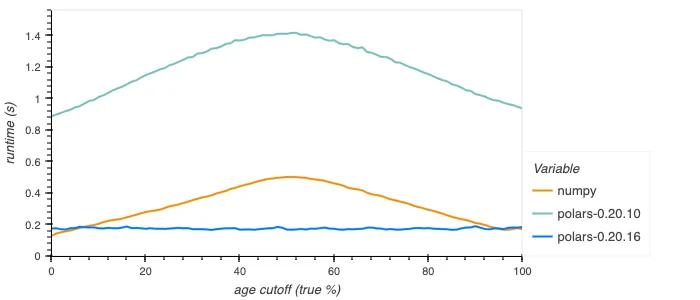
The old implementation was rather inefficient, going through dynamic dispatching internally for each element, as well as being potentially branchy. The new implementation is branchless and SIMD vectorized, giving constant good performance. For booleans the new implementation is much improved still, using efficient bitmask operations to perform the operation for 64+ elements at a time. Below is shown how this performs.
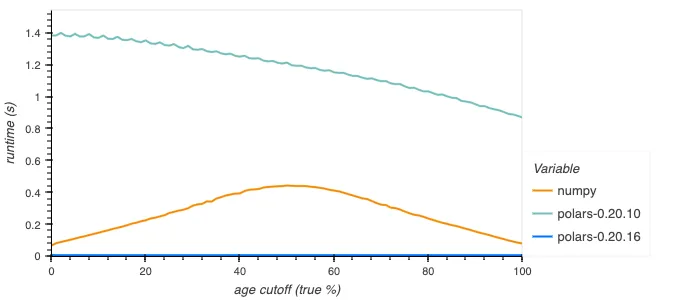
Parallelisation of sink_parquet (#14964)
Polars sink_parquet is of great use when dealing with very large datasets. Sinking to disk is a strategy that allows you to finish datasets that are larger than your available RAM size. This operation was still single threaded, leading to a lot of backpressure in the streaming engine. Now we fixed that bottleneck, greatly improving sinking performance.
Enchanced zero-copy interop with NumPy (#14434)
A lot of Python libraries rely on NumPy for efficient matrix computations. This is the case for many libraries in the machine learning space, including for example scikit-learn. In order to improve the integration of Polars with these libraries, we made a push to improve the performance of converting to and from NumPy arrays. We have improved our engine to reduce data copy to a minimum. That is, when we copy it actually benefits the query as future memory accesses will be cache efficient.
Differences in memory layout
There are a number of key differences between Polars and NumPy in the way data is represented in memory. These come into play when converting between the data formats in the most efficient way possible:
- Row-based representation: For two-dimensional data, NumPy arrays are typically represented in c-order which is row-oriented. Polars uses a columnar memory representation.
- Missing data: NumPy doesn’t have notion of missing data. And pandas made the decision to (mis)use NaN for this goal. Polars therefore follows this strategy when exporting to NumPy to follow the principle of least surprise. Internally Polars has validity buffers and therefore has a clear distinction between missing data and floating point Not A Number.
- Byte packed booleans: NumPy represents booleans as a byte (unsigned 8-bit integer that can be either 0 or 1), while Polars uses bitpacking to represent boolean values (8 booleans per byte).
- 64-bit dates: NumPy represents dates as a 64-bit datetime type with ‘day’ granularity, while Polars uses a 32-bit representation.
Even within these constraints, there are many cases where we can actually perform zero copy conversions. And even when zero copy conversion is not possible, there is some room for minimizing the amount of copy needed.
Improvements
Dealing optimally with the constraints outlined above required some effort, and we’re not completely done yet, but we’re happy with the recent improvements we made to our engine:
- Converting a 2D array from NumPy to Polars may now be zero-copy if the original array is already in Fortran (columnar) memory order.
- Converting a DataFrame from Polars to NumPy may now be zero-copy if the Series were all contiguous in memory.
- Some conversions had unnecessary data copy in certain situations. This has been cleaned up.
- Added parameters
allow_copy/writabletoDataFrame.to_numpyto control if/when data copy happens. UpdatedSeries.to_numpyto correctly respect these parameters in all cases.
Note that, at the time of writing, the default engine is still the PyArrow engine. To take advantage of these improvements, set use_pyarrow=False in your to_numpy call.
Next steps
One thing we still need to implement is the correct handling of nested data. There are some design decisions that must be made here. For example, do we automatically convert a Struct to a structured array, or do we simply convert to a NumPy object array?
We may also support converting to masked arrays in the future. This memory layout has a separate validity buffer, which more closely matches the Arrow layout. While the validity buffer will still need to be converted from bit packed to byte packed booleans, the data values would no longer need to be copied.
We aim to change the default engine for converting into NumPy from PyArrow to our own engine in the next breaking release.
Fast path for COUNT(*) queries (#14574)
The query SELECT COUNT(*) FROM SOURCE; (or source.select(pl.len()) in Polars syntax) is probably the most familiar query there is. It counts the number of rows in a data set, which you use to get a sense of the dimensions you are working with. Previously, Polars would materialize. It would read and parse a single column of the data set and count the number of rows in that column. This is sub-optimal because it takes time to deserialize and parse even a single column. With our new release, we improve this by using the underlying file format. In Parquet and Arrow, the number of rows is often stored in the metadata, eliminating the need to read any data at all. In CSV, we can simply count the number of rows in the file. Overall, this will significantly improve performance and makes memory usage negligable.
Proper handling of NaN, -0.0 and 0.0. in groupby and joins. (#14617)
In IEEE-754 float there are two representations for 0, as well as multiple possible representations of NaNs. This can cause problems in (hash-based) groupbys and joins where values that are logically equivalent end up in different groups. To prevent this we’ve adapted our internal hash and equality operators to normalize signed zero to positive zero and all NaNs to one with the same (bitwise) representation.
Community improved decimal support (#14209,#14338, #15000, #15001)
Decimal support has been improved a lot by community additions. We think the decimal support is now on a level we can it activate it by default in the upcoming Polars 1.0.
Integrations with the tools you use
In the past few weeks we have seen many new integrations with Polars. These integrations enrich the ecosystem so that you don’t have to convert a Polars dataframes to other libraries anymore. Below you can find four examples (in alphabetic order):
Great Tables
With Great Tables anyone can make wonderful-looking tables in Python. The philosophy here is that we can construct a wide variety of useful tables by working with a cohesive set of table components.
Read about Polars and Great Tables here
Toggle for code example
import polars as pl
from great_tables import GT, from_column, style, loc
from great_tables.data import airquality
airquality_mini = pl.from_pandas(airquality.head())
fill_color_temp = (
pl.when(pl.col("Temp") > 70)
.then(pl.lit("lightyellow"))
.otherwise(pl.lit("lightblue"))
)
(
GT(airquality_mini)
.tab_style(
style=style.fill(color=fill_color_temp),
locations=loc.body("Temp")
)
)| Ozone | Solar_R | Wind | Temp | Month | Day |
|---|---|---|---|---|---|
| 41.0 | 190.0 | 7.4 | 67 | 5 | 1 |
| 36.0 | 118.0 | 8.0 | 72 | 5 | 2 |
| 12.0 | 149.0 | 12.6 | 74 | 5 | 3 |
| 18.0 | 313.0 | 11.5 | 62 | 5 | 4 |
| None | None | 14.3 | 56 | 5 | 5 |
Hugging Face
Hugging Face offers a platform for the machine learning community to collaborate on models, datasets and applications. It is home to many open source models and datasets. Hugging Face added Polars support, so you can explore all datasets using the syntax you love.
Find out more about the dataset server on Hugging Face
Toggle for code example
import polars as pl
df = (
pl.read_parquet("https://huggingface.co/datasets/blog_authorship_corpus/resolve/refs%2Fconvert%2Fparquet/blog_authorship_corpus/train/0000.parquet")
.groupby("horoscope")
.agg(
[
pl.count(),
pl.col("text").str.n_chars().mean().alias("avg_blog_length")
]
)
.sort("avg_blog_length", descending=True)
.limit(5)
)
dfshape: (5, 3)
┌───────────┬───────┬─────────────────┐
│ horoscope ┆ count ┆ avg_blog_length │
│ --- ┆ --- ┆ --- │
│ str ┆ u32 ┆ f64 │
╞═══════════╪═══════╪═════════════════╡
│ Aquarius ┆ 34062 ┆ 1129.218836 │
│ Cancer ┆ 41509 ┆ 1098.366812 │
│ Capricorn ┆ 33961 ┆ 1073.2002 │
│ Libra ┆ 40302 ┆ 1072.071833 │
│ Leo ┆ 40587 ┆ 1064.053687 │
└───────────┴───────┴─────────────────┘Pandera
pandera is a Union.ai open source project that provides a flexible and expressive API for performing data validation on dataframe-like objects to make data processing pipelines more readable and robust.
Find out more about the integration in the documentation of Pandera.
Toggle for code example
import pandera.polars as pa
import polars as pl
class Schema(pa.DataFrameModel):
state: str
city: str
price: int = pa.Field(in_range={"min_value": 5, "max_value": 20})
lf = pl.LazyFrame(
{
'state': ['FL','FL','FL','CA','CA','CA'],
'city': [
'Orlando',
'Miami',
'Tampa',
'San Francisco',
'Los Angeles',
'San Diego',
],
'price': [8, 12, 10, 16, 20, 18],
}
)
print(Schema.validate(lf).collect())scikit-learn
Probably doesn’t need a lot of introduction. It is one of the most popular machine learning libraries. Many Polars users have requested a better integration. We are happy to share that transformers now support Polars output in scikit-Learn version 1.4.0 with set_output(transform="polars").
Toggle for code example
import polars as pl
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
df = pl.DataFrame(
{"height": [120, 140, 150, 110, 100], "pet": ["dog", "cat", "dog", "cat", "cat"]}
)
preprocessor = ColumnTransformer(
[
("numerical", StandardScaler(), ["height"]),
("categorical", OneHotEncoder(sparse_output=False), ["pet"]),
],
verbose_feature_names_out=False,
)
preprocessor.set_output(transform="polars")
df_out = preprocessor.fit_transform(df)
df_outshape: (5, 3)
┌───────────┬─────────┬─────────┐
│ height ┆ pet_cat ┆ pet_dog │
│ --- ┆ --- ┆ --- │
│ f64 ┆ f64 ┆ f64 │
╞═══════════╪═════════╪═════════╡
│ -0.215666 ┆ 0.0 ┆ 1.0 │
│ 0.862662 ┆ 1.0 ┆ 0.0 │
│ 1.401826 ┆ 0.0 ┆ 1.0 │
│ -0.754829 ┆ 1.0 ┆ 0.0 │
│ -1.293993 ┆ 1.0 ┆ 0.0 │
└───────────┴─────────┴─────────┘Final remarks
If you are in the area, join Ritchie’s keynote talk at PyCon Lithuania in Vilnius on 5 April 2024. Marco Gorrelli, Polars core dev, will be there to give a talk about DataFrame interoperatiblity and Luca Baggi will tell more about functime, a machine learning library for time-series predictions based on Polars.
On 2 April, Marco will also give a 4 hour deep dive tutorial to supercharge your data workflows with Polars. This PyCon is a great moment to connect, boost your Polars skill set and get your hands on some official Polars stickers.
We are hiring
We are hiring Rust software engineers. Do you want to contribute to one of the next Polars In Aggregates. See our jobs page for more info.